-How
does NO binding
-Peptide mimics
of the A-helix: Potential inhibitors of Hb S fiber formation?
Can
we monitor sickle cell polymerization in real time using UV Resonance
Raman (UVRR)?
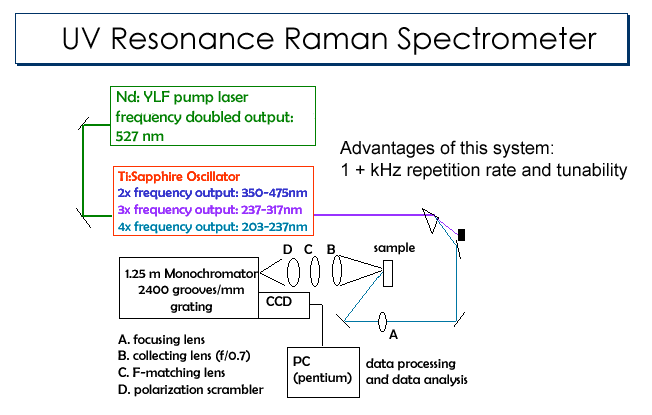
We
are currently modifying our UVRR spectrometer (FIG 10) to
have time-resolved capabilities. By
performing this modification, we will be able to acquire UVRR spectra as a
function of time and in this way monitor the changes in the Hb S tetramers
as they form fibers. In
particular we are very interested in monitoring the pre-gelation
aggregates, in other words those aggregates that form prior to the actual
fiber formation. These
aggregates form predominately during the 'delay time' period or homogenous
nucleation (link to that section). In
terms of the mechanism of fiber formation, this is one of the most
important steps.
|
|
| FIG 10 UV Resonance Raman Spectrometer |
How
does NO binding
Recent reports in the literature suggest that NO binding to Hb S can inhibit fiber formation and leads to the dissolution of fibers (link to Stamler lab on Nitrosylation: The Prototype Redox-Based Signal). One hypothesis is that NO binds preferentially to the b93 Cys residues in the R-state and deoxygenation of the hemes leads to a release of NO. Others have reported that hemoglobin is a scavenger of NO and that cell-free hemoglobin leads to sickle cell crisis because of this scavenging activity. (link to latest news stories re: NO) In our laboratory, we are probing the mechanism of action by investigating the affect of NO binding to the b93 Cys residues. In our experiments, the Cys residues are chemically modified to simulate the binding of NO. The polymerization potential of these modified adducts are being studied by kinetic assays and solubility studies. The structure of the fibers formed by Hb S molecules with modified cysteine residues will be studied using UV resonance Raman spectroscopy. These measurements will probe the intra- and intermolecular contacts of the fibers to determine if they are perturbed by the presence of the chemical modification at the b93 Cys position.
Peptide mimics of the A-helix: Potential inhibitors of Hb S fiber formation?
We are studying the action of several different
peptides on Hb S fiber formation. These
peptides, which are the same sequence as the A-helix of Hb S and contain
the b6
Val®Glu
mutation, can potentially interact with the hydrophobic EF corner and
prevent the interaction of other Hb S tetramers; thereby preventing fiber
formation. We have studied a
peptide, which consists of the first 8 residues of the A-helix and two 18
residue peptides, which comprise the entire length of the A-helix.
Our initial measurements indicated that the 18 residue peptide
potentially inhibits fiber formation.
Most recently, we have been trying to determine if the folded
state (link to protein structure/folding) of the peptide will
influence its inhibitory properties. We have determined that both the 8 residue and 18 residue
peptides fold in the presence of the solvent, trifluoroethanol (TFE).
Unfortunately, this solvent causes Hb S to precipitate therefore,
we cannot test the inhibitory properties under the conditions in which the
peptides are folded. We are
now trying to find other agents, such as dextran, that might induce the
peptides to fold.
Back to top